Goal:
During the tutorial, we will learn how to screen chemical libraries against toxicological alerts, to identify potentially hazardous compounds and to interpret the results. For accessing this handout and other related materials, please visit http://docs.eadmet.com/display/EWS2M/
Motivation:
- Do you have a practically interesting set of molecules? Screen them with a few clicks, analyze the results and publish your screening results in a scientific article.
- Have a set of your own alerts? Share them with the community and get cited.
Datasets:
For speed and convenience, we will not use any external SD-files. We have already uploaded the necessary datasets into OCHEM.
Tutorial 1 – Basic screening for several endpoints
In this tutorial we will learn how to browse alerts, create alert sets, screen chemical libraries against the desired alerts and analyze the results.
Datasets.
We will use the "High performance volume" dataset. The whole dataset has been split into "HPV low mol. Weight" and "HPV high mol. weight" according to the molecular weight of the structures. Please, use any of the datasets.
1. Choosing the desired alerts
We would like to screen our molecules against five endpoints: acute aquatic toxicity, skin sensitization, non-genotoxic carcinogenicity, genotoxic carcinogenicity and idiosyncratic toxicity.
- To select the relevant alerts, go to the alerts browser (Menu item: Database ->ToxAlerts -> View available alerts)
- Specify the filter for endpoint "acute aquatic toxicity" (99 alerts should be filtered)
- Save these alerts to your set by clicking "select all" icon:

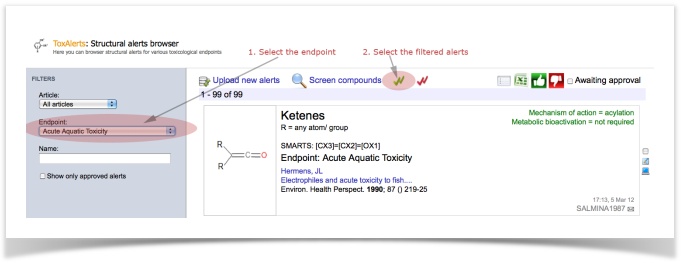
- Repeat the same steps the other endpoints: skin sensitization (161 alerts), non-genotoxic carcinogenicity (5 alerts), genotoxic carcinogenicity (117 alerts) and idiosyncratic toxicity (35 alerts).
- In the end, you should have selected 417 alerts.
Now, we are ready to screen molecules against the selected alerts!
2. Screening molecules against alerts
- Open the screening dialog (Menu item: Database – ToxAlerts - Screen compounds against alerts)
- Select the desired set (e.g., molecules by tag, the "HPV low mol. weight" dataset)
- Select the desired alerts (remember, we have selected 417 alerts for five endpoints? Check the "only 417 selected alerts")
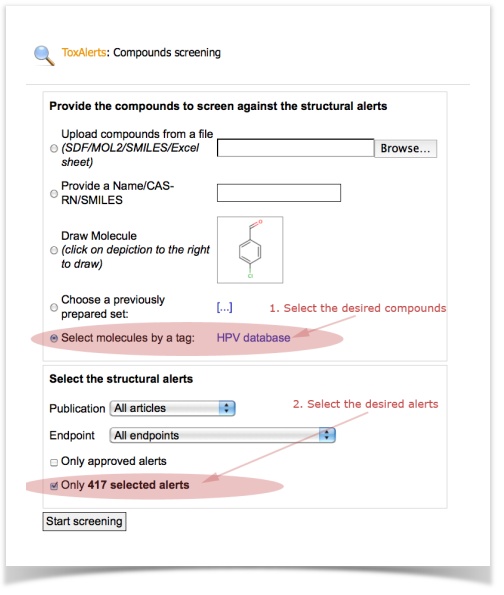
- Start screening and wait for the calculation task to finish. Can take 1-5 minutes for the set with several thousands molecules.
- You can get back to the task any time later in the browser of pending tasks (Menu item: Models – View pending tasks).
3. Analyze the screening results
The screening results dialog shows the potentially hazardous compounds grouped by endpoints, by alerts and by publications. Please, spend some time to click around.
- View the compounds for a particular endpoint (e.g., non-genotoxic carcinogenicity)

- Tag the filtered compound (e.g., "HPV non-genotoxic carcinogens")
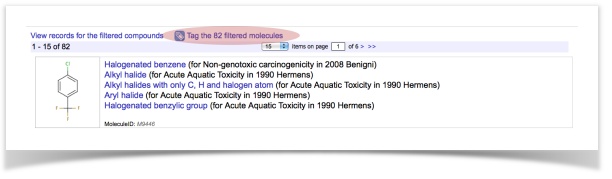
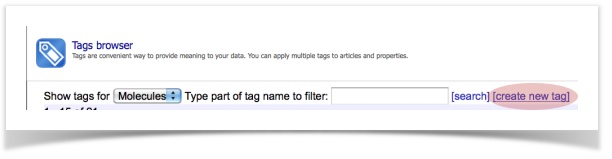
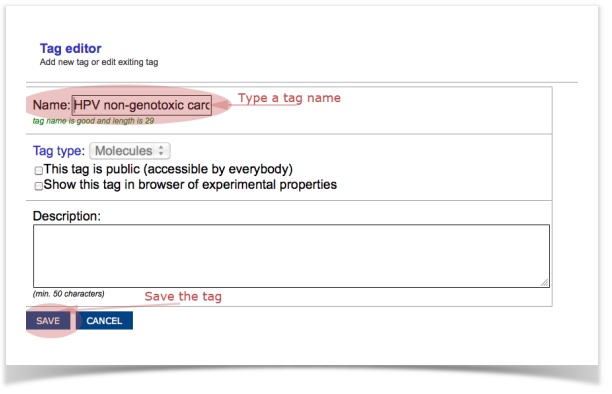
- Access, view or export the tagged structures into an SD-file via the browser of tags (Menu item: Database – Tags)
Tutorial 2 – SetCompare using functional groups
In this tutorial, we will use a dataset of ready-biodegradability. We have prepared two sets of molecules containing (a) 717 readily biodegradable compounds and (b) 1221 non-readily biodegradable compounds.
The datasets were taken from the study of Vorberg et. Al "Modeling the biodegradability of chemical compounds using the On-line CHEmical modeling Environment (OCHEM)", J. Mol. Inf, in review.
Follow the steps below:
- Make sure you have the datasets available (Menu item: Database – Tags, activate the name filter by typing "ECO")
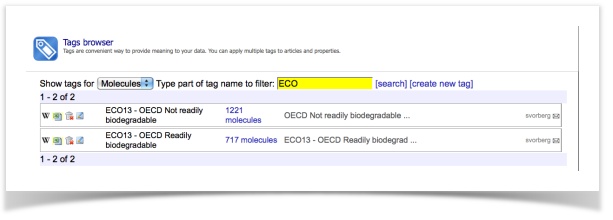
- Open the SetCompare utility (Menu item: Models – SetCompare utility)
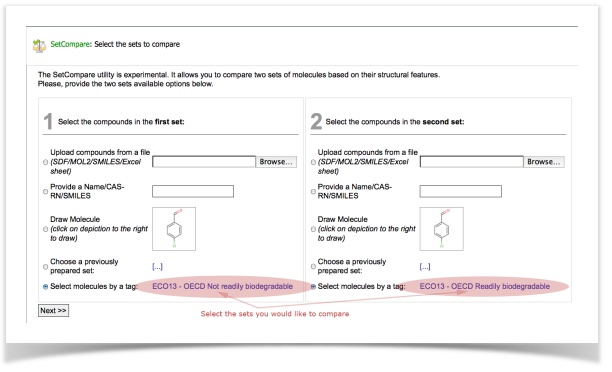
- Select the tags with readily- and non-readily biodegradable compounds and click "Next"
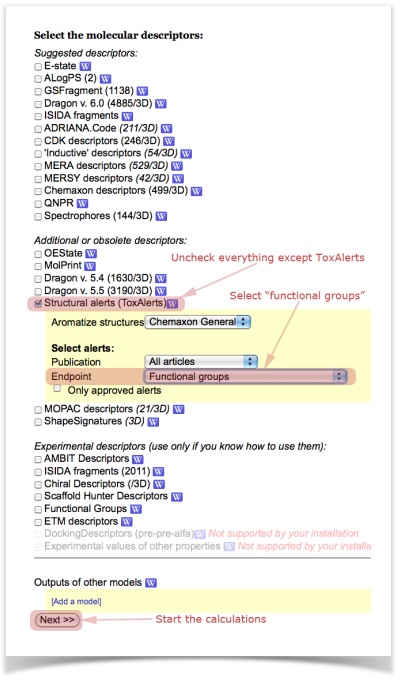
- Now we have to choose the descriptors used to compare our sets. W will use ToxAlerts' functional groups. Make sure you uncheck all descriptor types except of "Structural alerts". Select the "functional groups" in the endpoint filter.
- Again, you can access your calculation task any time later from the browser of pending tasks (Menu item: Models – View pending tasks)
Finally, when your calculation is completed, review and analyze the results!
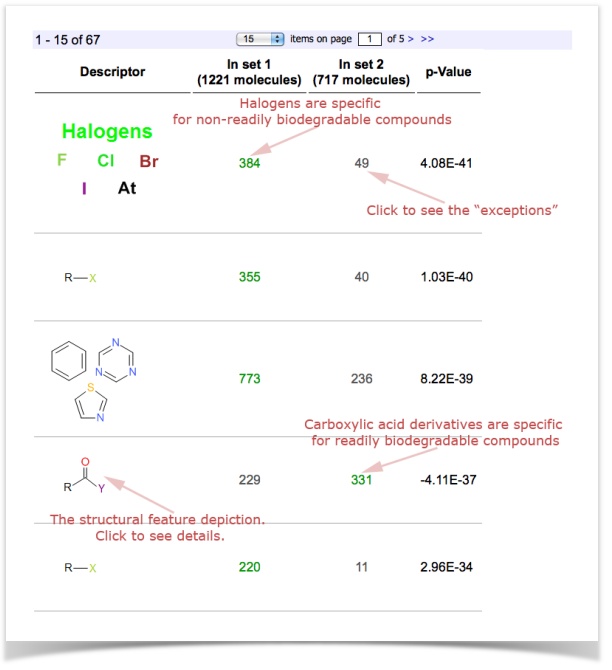
The screen with results will show you the structural features that are statistically over-represented in the one or the other set.
This exemplary study shows you, for example, that Halogens are over-represented in non-readily biodegradable compounds.
Conclusion
The tutorials demonstrate a fundamental difference of ToxAlerts (and the alerts-based analysis) from traditional QSAR: interpretability of the former approach.
While QSAR approaches are, in general, more powerful, universal and accurate, the alerts-based approach allows to obtain more interpretable results.
Thus, alerts-based analysis is a useful utility in the toolbox of a chemoinformatician that can complement the conventional QSAR techniques.