MolPrint (aka MolPrint 2D) descriptors[1,2] are a particular type of circular fingerprint which employ Sybyl MOL2 atom types. More specifically, they are based on counts of MOL2 atom types around each heavy atom of the molecule. In contrast to structural keys they do not draw features from a limited set of structural fragments (such as MACCS keys). Rather, they enumerate all atom environments present in a molecule. MolPrint 2D descriptors are similar to SciTegic's (Pipeline Pilot) extended-connectivity fingerprints (ECFP), but MolPrint 2D features are not hashed.[5] The implementation of MolPrint 2D used in OCHEM uses the atom types literally as they appear in a MOL2 file, i.e., an aromatic carbon is encoded as "C.ar", a sp2-hybridized oxygen atom as "O.2", etc.
For each heavy atom all neighboring atoms at a given number of bonds away are tallied and encoded as a string. Such a string always starts with the heavy atom C at the center of the feature, followed by triples of the form D-T-N, where D is the distance in bonds from the central atom (D in {1, 2, …}), T the type of atom (T is a valid Sybyl MOL2 atom type), and N the number of atoms of type T that can be found at a distance D of the central atom C. The central atom and all tripled are separated by semicolons. Overall, that results in feature strings of the form: C;D-T-N;D-T-N;D-T-N;… In practice, it was found that values for D up to 3 should be considered for descriptor generation, with D=2 the most commonly employed. The higher the value for D, the more specific the features become by nature of their construction.
A feature that would be generated for the atom marked in bold in this figure (the central atom for this feature)
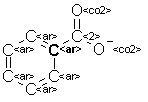
would be described as follows:
Central atom: C.ar
Distance of one bond from C: two times C.ar => 1-C.ar-2; one timee C.co2 => 1-C.co2-1;
Distance of two bonds from C: two times O.co2 => 2-O.co2-2; two times C.ar => 2-C.ar-2;
The final feature for the above example would be the concatenation of the central atom and all the triples: C.ar;1-C.ar-2;1-C.co2-1;2-C.ar-2;2-O.co2-2;
For each distance D, the triples are ordered alphabetically, so 1-C.ar-2 would come before 1-F-2 but after 1-Br-1. In the example above, 2-C.ar-2 comes before 2-O.co2-2.
This procedure is repeated for every heavy atom in the molecule.
The binary nature of descriptors renders MolPrint descriptors more amenable to certain types of modeling methods (such as Bayes or k-NN methods), more than for example neural network models. The models generated are relatively easy to interpret, since every feature corresponds to roughly a functional group (though without explicit information about the bond order between atoms).
MolPrint descriptors have been used successfully in virtual screening[3] and ligand-target prediction[4] where they have been shown to capture a large amound of the information relating molecular structure to bioactivity against a protein target.
References
[1] A. Bender, H.Y. Mussa, R.C. Glen and S. Reiling. Molecular similarity searching using atom environments, information-based feature selection, and a naive bayesian classifier. Journal of Chemical Information and Computer Sciences, 2004, 44, 170-178. - http://dx.doi.org/10.1021/ci034207y
[2] A. Bender, H.Y. Mussa, R.C. Glen and S. Reiling. Similarity searching of chemical databases using atom environment descriptors: evaluation of performance. Journal of Chemical Information and Computer Sciences, 2004, 44, 1708-1718. - http://dx.doi.org/10.1021/ci0498719
[3] R.C. Glen, A. Bender, C.H. Arnby, L. Carlsson, S. Boyer and J. Smith. Circular fingerprints: Flexible molecular descriptors with applications from physical chemistry to ADME. IDrugs 2006, 9, 199-204. - http://www.biomedcentral.com/content/pdf/cd-653859.pdf
[4] Nidhi, M. Glick, J. W. Davies and J. L. Jenkins. Prediction of Biological Targets for Compounds Using Multiple-Category Bayesian Models Trained on Chemogenomics Databases. J. Chem. Inf. Model., 2006, 46, 1124–1133. - http://pubs.acs.org/doi/abs/10.1021/ci060003g
[5] Rogers and Hahn. Extended-Connectivity Fingerprints. J Chem Inf Model. 2010 May 24;50(5):742-54.