Molecular descriptors are numerical features of chemical compounds.
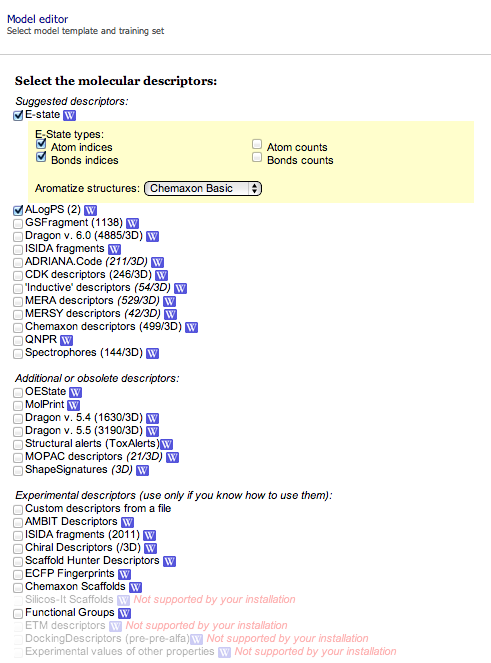
OCHEM supports the calculation of many different descriptor packages:
- Adriana.CODE
- alvaDesc
- ALogPS
- CDK
- Chemaxon descriptors
- Chemaxon scaffolds
- Custom descriptors from a file
- Dragon
- ECFP fingerprints
- ESTATE
- Experimental values of other properties
- Functional groups
- GSFrag
- Inductive Descriptors
- ISIDA Fragments
- MERA descriptors
- MERSY descriptors
- MolPrint
- MOPAC-derived descriptors
- OEState
- QNPR
- Silicos-It scaffolds
- Spectrophores
- ToxAlerts
- MAP4
- JPLOGP
- RDKit descriptors
- MOE
- CDDD
- PyDescriptor
- CDK22
- SIRMS
- CDK2
- RDKit additional descriptors
- MORDRED
- MOPAC2016-derived descriptors
You can select one or more descriptor packages to calculate for the same dataset. The calculation time for descriptors depends on many factors including the size of the dataset, the package selected, and the availability of computational resources.
Page: Adriana.CODE
Page: alvaDesc
Page: ALogPS
Page: CDK
Page: Chemaxon descriptors
Page: Chemaxon scaffolds
Page: Custom descriptors from a file
Page: Dragon
Page: ECFP fingerprints
Page: ESTATE
Page: Experimental values of other properties
Page: Functional groups
Page: GSFrag
Page: Inductive Descriptors
Page: ISIDA Fragments
Page: MERA descriptors
Page: MERSY descriptors
Page: MolPrint
Page: MOPAC-derived descriptors
Page: OEState
Page: QNPR
Page: Silicos-It scaffolds
Page: Spectrophores
Page: ToxAlerts
Page: MAP4
Page: JPLOGP
Page: RDKit descriptors
Page: MOE
Page: CDDD
Page: PyDescriptor
Page: CDK22
Page: SIRMS
Page: CDK2
Page: RDKit additional descriptors
Page: MORDRED
Page: MOPAC2016-derived descriptors