Introduction
A molecular matched pair (MMP) is a pair of molecules that have only a minor single-point difference.
The typical way is to define a minor difference as a changed molecular fragment with less than 10 atoms.
Examplary MMPs (differences highlighted in orange):
Why MMPs?
MMPs aid interpreting how particular structural changes affect the chemical and biological properties of a molecule. For example, how addition or replacement of a particular funtional group affects the solubility, melting point or toxicity of a molecule?
Definitely, such interpretation is possible only If the experimental data (or predicted values) is available for both molecules in the pair.
For example, in the MMP below, a minor change resulted into molecule being active according to the Ames test:
In the following example, after a minor structural change, a molecule changed its inhibitory activity for the human P450 cytochrome :
MMPs and OCHEM
Automatic identification of MMPs
OCHEM has a separate module that allows to identify molecular matched pairs, to group them by molecular transformations and to calculate various statistical information for particular pairs or transformations.OCHEM automatically indexes all the molecules having any experimental data associated with them and identifies the matched pairs.
The pairs identification process is running in background and can take some time (hours to days) for new molecules.
Matched pairs and transformations
Each matched molecular pair is related to a particular molecular transformation. For example, replacement of a functional group with another group is a transformation. OCHEM groups all pairs by transformations allows to see them in the transformations browser, as show in the screenshot below.
Each transformation has its profile page that contains different details: SMIRKS of the transformation, all the molecular pairs related to this transformation and, if applicable, affected properties and activities:
Transformations annotation
Transformations can affect particular properties. For example, alter toxicity, lipophilicity, melting point and so on.
OCHEM can automatically identify and annotate transformations that affect various molecular properties in a statistically significant manner.
Such annotations is shown on the transformations profile.
For each affected property, it is possible to see a basic statistical info, a histogram of the pair deltas and all the pairs used to calculate statistics for this transformation.
To select the transformations that affect the desired properties or activities, there is a transformation optimiser utility.
The following screenshot shows the transformations that affect the inhibition growth concentration (log[ICG50]) but do not affect lipophilicity:
MMP-based interpretation of QSAR models
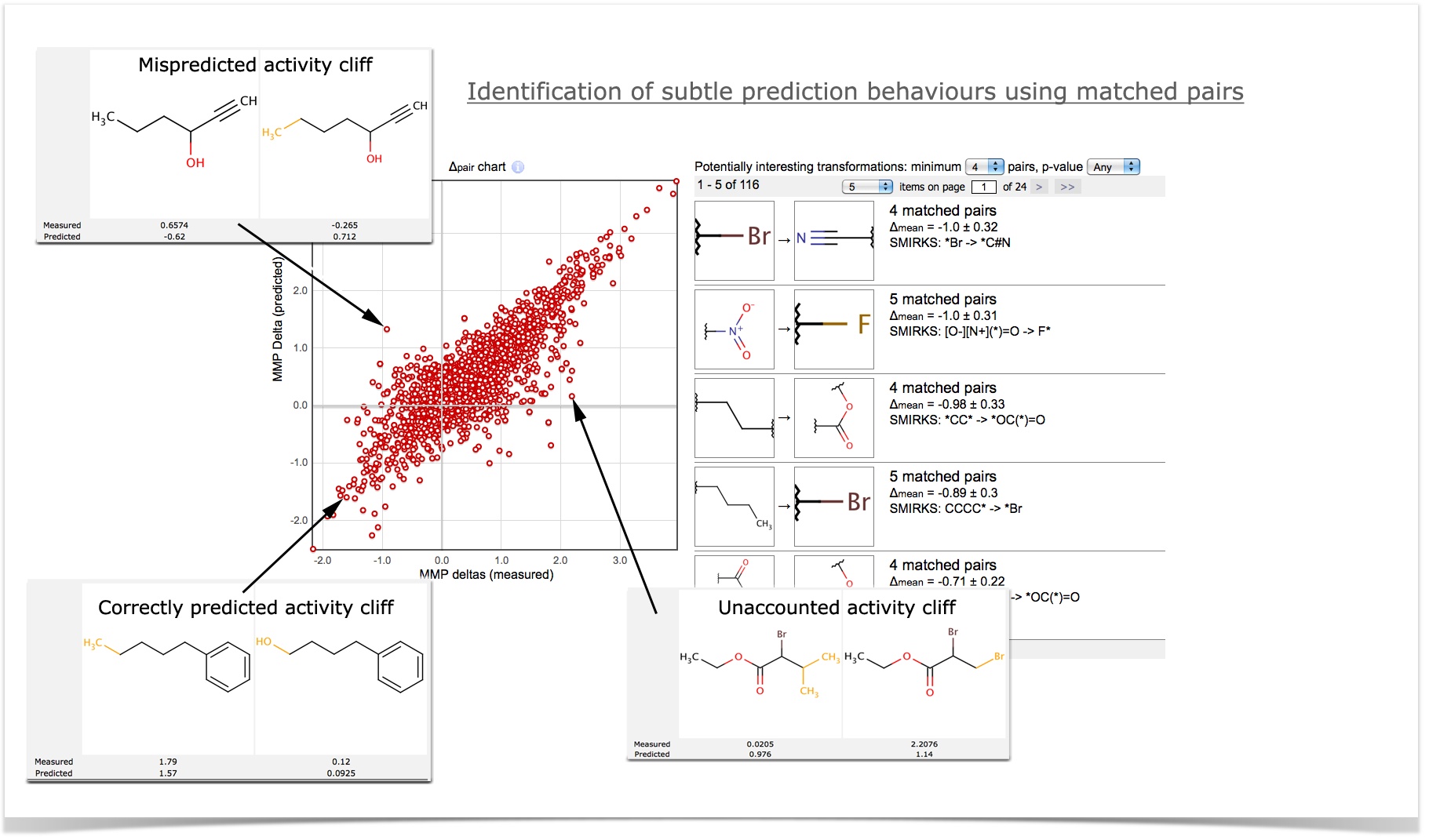
MMPs can be used to identify subtle prediction behaviours, e.g., model reaction to activity cliffs. The delta-pair chart shows the actual and predicted effects of molecular transformations.
Significant molecular transformations. The list on the right shows the "interesting" molecular transformations that have a statistically significant effect on the property of interest. From the screenshot below, it can be seen that replacement of Bromine to Nitrile group (1st transfomation in the list) reduces toxicity of compounds.
We will call such transformations "significant".
Molecular Fragments Graph
Each molecular transformation is a replacement of one molecular fragment to another one. That is, a transformation is a relation between two molecular fragments.
Based on the "significant" transformations, it is possible to create a directed graph of molecular fragments. Each node in this graph is a fragment, each edge is a significant transformation. Such graphs can display a number of transformations and allow for a better interpretation.
The following graph display a part of the transformations related to Aquatic toxicity. Arrows point the direction of increasing toxicity. For example, it can be seen that the presence of Bromine is potentially more toxic than the presence of Chlorine.
References
The article with description of a use of MMPs to analyze models has been published in Prediction-driven matched molecular pairs to interpret QSARs and aid the molecular optimization process. Sushko Y, Novotarskyi S, Körner R, Vogt J, Abdelaziz A, Tetko IV. J Cheminform. 2014 Dec 11;6(1):48. doi: 10.1186/s13321-014-0048-0.









