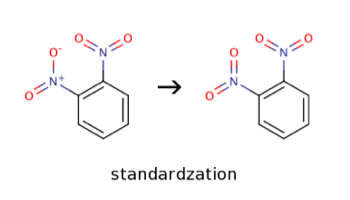
Standardization
Standardization is the process of transforming a molecule according to a set of SMARTS templates. The templates currently used in OCHEM allow converting nitro mesomers. It is a required step to receive consistent molecule datasets. Due to limitations of molecule representation in QSAR, molecules with different nitro mesomer representations may be treated as different molecules. This is wrong from a chemical and biological point of view. The following image reflects a sample transformation of a molecule if standardization is enabled.
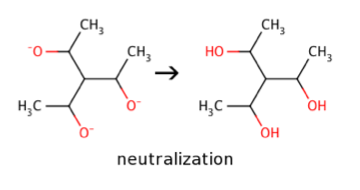
Neutralize
Neutralization refers to neutralization of charged atoms in the molecules by attaching additional hydrogen atoms to them. Mesomers like nitro groups or quaternary nitrogens without hydrogens remain intact.
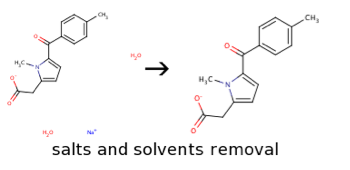
Remove salts
Remove salts is a procedure that allows removing salts, counter-ions, solvents and other molecule fragments from molecular structure. From all the detached fragments the biggest by mass is kept. It is an important step, since a large amount of molecule optimization or molecule descriptor calculation tools can not correctly process molecules containing salt or counter-ions. This procedure, however, results to loss of information on complete molecule structure and may lead to false duplicates in analyzed datasets.
Clean structure
Clean structure is a procedure where the original molecule file is converted to SMILES format and back, which results into complete loss of all information in a molecule except atom connectivity. This is useful to remove any 3D or atom coordinate calculation information, which in a number of cases has been show to cause model overfitting.
For a more detailed description of Chemaxon Standardizer preprocessing options, please refer to the official Chemaxon documentation.