OCHEM allows creating multiple models with different configuration options. Hundreds of models can be created with a few clicks.
The utility for multiple models creation and management is referred to as "comprehensive modeling" utility.
Creation of multiple models
To access the comprehensive modelling dialogue, select the menu "Models > Create multiple models".
Select the necessary training and validation sets.
Select the predefined configuration templates for:
- molecular descriptors
- machine learning methods
- descriptors filtering
- validation protocols
Each combination of the selected templates will be used, therefore, if you select 2 types of descriptors, 3 machine learning methods, 1 descriptor filtering options and 2 validation options, then 2x3x1x2 = 12 models will be created.
Wait until the models have been started:
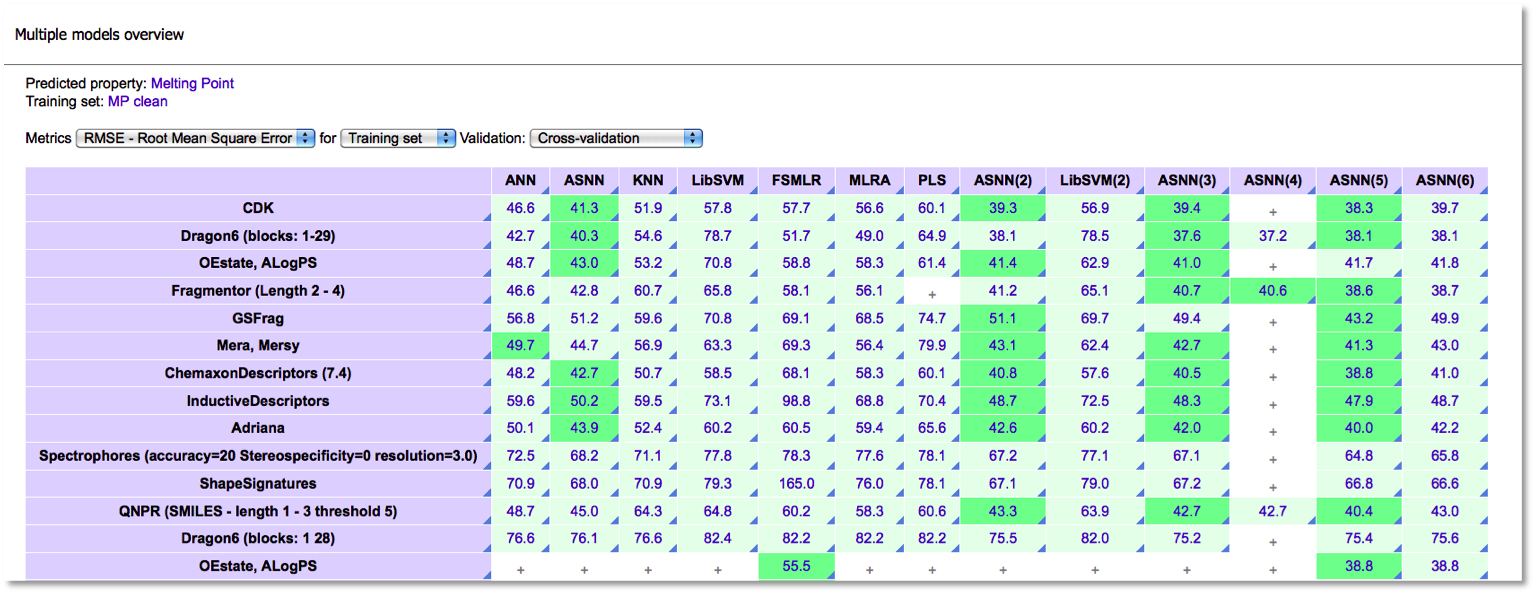
Multiple models overview
The multiple models overview page will display the status of all the models, both running and completed:
When the models are ready, we fetch the statistics by clicking the appropriate lick and overview the predictive performance of the models.
The multiple models overview allows to compare models by:
- RMSE
- R2
- Mean absolute error (MAE)
It is possible to compare the models by their performances for both training and validation sets.
The screenshot below displays RMSE values for 9 models developed for BCF (bio-concentration factor) endpoint based on the "BCF train" basket.
Comprehensive modeling can be a very powerful feature. Which descriptors are the best? How do the models evolve when outliers are excluded? Which training method performs best for this property? All this questions require deep analysis made possible using comprehensive modeling.
The screenshot below shows an intermediate result of a real on-going study – prediction of melting point based on more than 30,000 experimental measurements. More than 150 models have been built. Using the comprehensive modeling feature, it was possible to identify the best methods and to gradually improve models by removing outliers and reducing noise.